Photo: Ksergent527 CC BY-SA4.0
PUBLICATIONS
2021
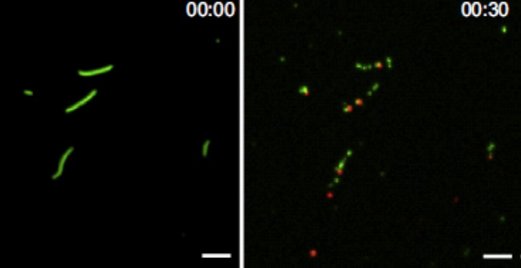
Figure: Images obtained using TIRF microscopy showing actin filaments (green) bound to surface adsorbed myosin motor fragments before (left) and 30 s after (right) addition of 5 nM Alexa647-labelled (red) gelsolin in the presence of micromolar Ca2+. Note presence of single, red, gelsolin molecules at several filament ends after the gelsolin induced fragmentation, consistent with gelsolin capping of one of the filament ends after fragmentation.
Focus on Biocomputation: Materials, algorithms, devices and fabrication
This special issue in the New Journal of Physics presents articles on the progress in bio- and bio-inspired computation. Specifically, development of new materials and fabrication technologies, achieved in an interdisciplinary approach engaging materials science, computer science, biophysics, cognitive science, micro- and nanofabrication, sensing, electronics and photonics.
Keywords: biological computation, parallel computing, subset sum problem, molecular motor, network-based biocomputation, nanofabrication, NP-complete problem, exact cover, microfluidics, e-beam lithography, actin, myosin, in vitro motility assay, electrostatic detection, nanoscale biosensors, interference reflection microscopy, microtubules, kinesin, gliding assay;
During 2021, the collection has grown to 8 articles and gives a comprehensive overview of the field. All articles are fully open access.
Special Issue in the New Journal of Physics
Actomyosin in Biocomputation
PhD thesis by Aseem Salhotra, Linnaueus University.
Actin and myosin constitute one type of molecular motor system that has been utilized for the development of biocomputation. These proteins are key components in the sarcomere, the smallest functional unit of muscle and their interactions that underlie muscle contraction are powered by the cellular fuel adenosine triphosphate. To solve larger complex problems using actin-myosin based biocomputation, factors such as maintained biological function and longevity of operation are essential for practical relevance.
VMCAI 2021artifact evaluation badge to verification of biocomputation circuits
Aluf-Medina M., Korten T., Raviv A., Nicolau Jr. D.V., Kugler H. (2021) Formal Semantics and Verification of Network-Based Biocomputation Circuits. In: Henglein F., Shoham S., Vizel Y. (eds) Verification, Model Checking, and Abstract Interpretation. VMCAI 2021. Lecture Notes in Computer Science, vol 12597. Springer, Cham.
https://doi.org/10.1007/978-3-030-67067-2_21
Single molecule turnover of fluorescent ATP by myosin and actomyosin unveil elusive enzymatic mechanisms.
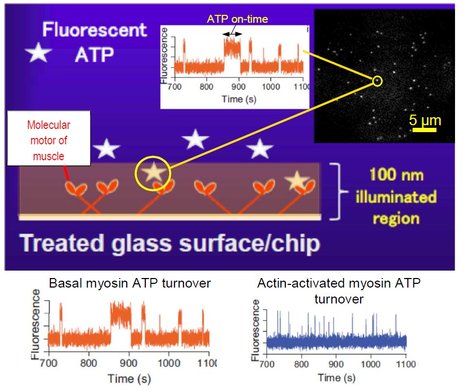
We have here developed a refined fluorescence microscopic method to visualize single molecules with negligible interference from fluorescent contaminants and complicated physical properties of fluorescent molecules such as photobleaching and photoblinking. These methodological advancements allow us to unveil previously hidden mechanisms in the ATP turnover by myosin and to pursue single molecule studies of the ATP turnover by myosin acting together with actin, a process appreciably faster than the myosin ATPase. Our approach utilizes, so called, total internal reflection fluorescence (TIRF) illumination (Fig. 1) to visualize only molecules very close (~100 nm or less) to the illuminated surface. Therefore, a fluorescent ATP molecule is seen in the microscope only when it is attached to surface-adsorbed enzymes, in our case, the molecular motor myosin of muscle. By analyzing the distribution of ATP-on-times for single molecules (bright dots in Fig. 1) we can measure the ATP turnover rate from one molecule at a time. The method is useful in Bio4comp as basis for the development of fluorescence detectors and for analysing the function of surface-adsorbed motors e.g. in a Biocomputation network. Additionally, the methods have been used in Bio4comp to study mechanisms in the multiplication of actin filaments, currently in press in the Journal of Biological Chemistry. More generally, the improvements will benefit the active field of single-molecule fluorescence studies beyond myosin and molecular motors. The work also paves the way for reliable single molecule analysis in high-throughput screening of drug candidates with minimal requirement of proteins from costly expression systems, individual cells or clinical samples with relevance from an ethics and sustainability perspective.
Figure: Principle of method, single ATP molecules (bright dots) bound to myosin motor fragments on a surface and recordings of fluorescence intensity vs time showing one-step on/off switching as ATP binds to/detaches from myosin.
Ušaj, M., Moretto, L., Vemula, V. et al. Single molecule turnover of fluorescent ATP by myosin and actomyosin unveil elusive enzymatic mechanisms. Commun Biol 4, 64 (2021). Open access;
https://doi.org/10.1038/s42003-020-01574-0
For certain designs of network-based biocomputation devices that make use of molecular motor propelled filaments, it is of importance to increase the number of filaments during their passage through the computation network. Such filament multiplication should be achievable by consecutive steps of filament splitting and elongation. One approach for effective filament splitting is studied in this paper, using the actin filament severing protein gelsolin. An important method in the study is the recently refined single molecule fluorescence microscopy assay based on total internal reflection fluorescence (TIRF) microscopy that we recently published in Communications Biology. In addition to the value for development of effective biocomputation approaches, the present study reveals phenomena of basic biophysical and cell physiological relevance. First, the results unveil novel aspects of cooperativity between gelsolin induced structural changes of the actin filaments and structural changes produced during myosin-induced production of force and motion. Second, we also provide arguments that these cooperative phenomena may be of great importance for the optimized control of cell function under specific conditions.
2019
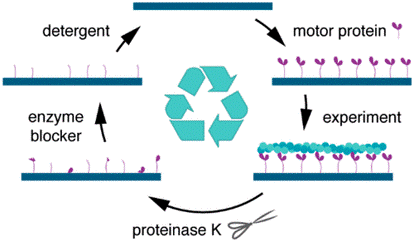
A new method to regenerate bionanodevices forms an important milestone to saving material and energy and thus, towards sustainable alternative chip technology. The devices do not need to be disassembled, but are treated with proteinase K and washed with mild detergent. Thereafter the surfaces can be functionalized with motility proteins again and used for subsequent gliding assays. The quality of the assays after regeneration is similar to before.
Regeneration of Assembled, Molecular-Motor-Based Bionanodevices
M.A. Rahman, et al. Nano Letters 2019
- This thesis shows that actin is a dynamic filament upon myosin binding and this information is useful when using actin filaments for Biocomputing.
- To control actin filament velocity myosin inhibitors, such as blebbistatin, can be useful for the in vitro motility assays in biocomputation network.
- Presence of dead myosin heads negatively affect the actin filament gliding in the in vitro motility assay used in a biocomputation network. This thesis describes and analyses two methods to remove the dead heads to provide good quality assay with nearly 100% fraction of motile filaments recovered.
- Biocomputation devices are expensive and time consuming to produce. This thesis describes a method where it is shown that the use of proteinase K with a suitable detergent can be useful to recycle nanodevices for several times.
- This thesis studied different ways to immobilize engineered light controlled myosin motor on TMCS derivatized surfaces that shed lights to develop a future programmable gate for a biocomputation network.
- Abstract: Biophysical studies of the actin-myosin motor system and applications in nanoscience. Mohammad Ashikur Rahman, 2019
Two important techological developments form the cornerstones of Frida Lindberg's doctoral thesis:
- the surface chemistry of the devices, which is altered in order to enable re-use and
- a more efficient fabrication method
Ups and downs of two standard ways to improve motility in the in vitro motility assay by removing the effects of "dead" non-functional heads.
Rahman, M.A., Salhotra, A. & Månsson, A. J. Comparative analysis of widely used methods to remove nonfunctional myosin heads for the in vitro motility assay. Muscle Res Cell Motil (2019).
https://doi.org/10.1007/s10974-019-09505-1
High throughput nanofabrication: To enable large scale network-based computation using molecular motors, a key requirement is the ability to fabricate big networks with a high resolution at a high throughput. All these devices require loading zones to feed filaments into the channel networks, regardless of the computational problem encoded. Nanoimprint lithography is an excellent, large scale fabrication method, but becomes difficult for high aspect ratio structures such as loading zones. By introducing nanoscaled pillars, we are able to high aspect ratio structures and provide a possibility to tailor the emptying rate and directional guiding of actin filaments.
Lindberg, F. W. et al. Design and development of nanoimprint-enabled structures for molecular motor devices. Materials Research Express 6, (2019).
2018
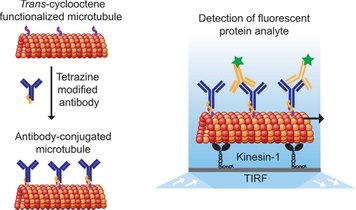
Development of miniaturized devices for the rapid and sensitive detection of analyte is crucial for various applications across healthcare, pharmaceutical, environmental, and other industries. Here, we report on the detection of unlabeled analyte by using fluorescently labeled, antibody-conjugated microtubules in a kinesin-1 gliding motility assay. The detection principle is based on the formation of fluorescent supramolecular assemblies of microtubule bundles and spools in the presence of multivalent analytes. We demonstrate the rapid, label-free detection of CD45+ microvesicles derived from leukemia cells.
Chaudhuri, S. et al. Label-Free Detection of Microvesicles and Proteins by the Bundling of Gliding Microtubules. Nano Letters 18, 117-123, 2018 doi:10.1021/acs.nanolett.7b03619
In order to facilitate rapid screening of compounds affecting kinesin motor activity as well as optimizing kinesin-based in vitro motility assays for nanotechnological applications, we developed an in vitro motility assay where sample preparation, imaging and data evaluation are fully automated, enabling the processing of a 384-well plate within less than three hours. We demonstrate the automated assay for the analysis of peptide inhibitors for kinesin-1 at a wide range of concentrations.
Source code for the automated data evaluation on GitHub
Korten, T., Tavkin, E., Scharrel, L., Kushwaha, V. S. & Diez, S. An automated in vitro motility assay for high-throughput studies of molecular motors. Lab on a Chip 18, 3196-3206, 2018.
doi:10.1039/c8lc00547h
Controlled surface chemistry: The guiding of actin filaments is strongly regulated by the surface chemistry. By regulating the surface hydrophobicity using only one type of silane, we are able to maintain the same, motility promoting chemical environment while tuning the actin filament velocity.
Lindberg, F. W. et al. Controlled Surface Silanization for Actin-Myosin and Biocompatibility of New Polymer Resists. Langmuir 34, 8777-8784, 2018.
doi:10.1021/acs.langmuir.8b01415
This is a review-article about the extent by which results from studies of single molecules of myosin and actin directly predict mechanical and contractile properties of muscle cells. The paper generally finds good agreement between model behavior based on parameters from single molecule studies and experimental data from muscle. This suggests that the 3D order of the contractile machinery in muscle together with a range of accessory proteins in addition to actin and myosin only have a minor modulatory role in the mechanisms leading to development of force and motion. This knowledge about basic mechanisms relies on input from Bio4comp but is also important for the project as actin and myosin are molecular work-horses that drive our computational machines.
Mansson, A., Usaj, M., Moretto, L. & Rassier, D. E. Do Actomyosin Single-Molecule Mechanics Data Predict Mechanics of Contracting Muscle? International Journal of Molecular Sciences 19, (2018).
doi:10.3390/ijms19071863
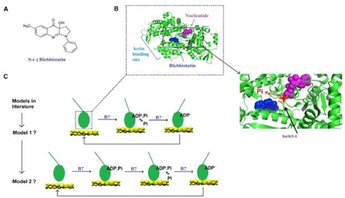
The small-molecular chemical compound blebbistatin can be used together with varied ATP concentration and varied ionic strength to fine-tune the gliding velocity of myosin propelled actin filaments in biocomputation devices. This is desirable e.g. to tune multiplication rate of actin filaments to velocity. In this study we characterize the effects of blebbistatin on velocity as well as its mechanism of action on the force- and motion-generating cycle of myosin with actin. In these studies we also unveil hidden secrets in this process in the absence of any modifying substance.
Rahman, M. A., Usaj, M., Rassier, D. E. & Mansson, A. Blebbistatin Effects Expose Hidden Secrets in the Force-Generating Cycle of Actin and Myosin. Biophysical Journal 115, 386-397, (2018).
doi:10.1016/j.bpj.2018.05.037
Verardo, D. et al. Nanowires for Biosensing: Lightguiding of Fluorescence as a Function of Diameter and Wavelength. Nano Letters 18, 4796-4802, (2018).
doi:10.1021/acs.nanolett.8b01360
2017
Engineering cargo-loading strategies is crucial to developing nanotechnological applications of microtubule-based biomolecular transport systems. Here, we report a highly efficient and robust bioconjugation scheme to load antibodies to microtubules.
Chaudhuri, S., Korten, T. & Diez, S. Tetrazine-trans-cyclooctene Mediated Conjugation of Antibodies to Microtubules Facilitates Subpicomolar Protein Detection. Bioconjugate Chemistry 28, 918-922, (2017).
doi:10.1021/acs.bioconjchem.7b00118
This review article describes the biological properties of actin filaments and how these properties may be altered by chemical engineering of the filaments. Such engineering is of relevance for Bio4Comp both for efforts to multiply actin filaments, to make them less flexible and to tag the filaments for certain computation approaches.
Kumar, S. & Mansson, A. Covalent and non-covalent chemical engineering of actin for biotechnological applications. Biotechnology Advances 35, 867-888, (2017).
doi:10.1016/j.biotechadv.2017.08.002
Highly-efficient guiding of motile microtubules on non-topographical motor patterns. A UV-laser-based ablation technique can be used to programmably generate highly localized patterns of functional kinesin-1 motors on PLL-g-PEG-coated, planar polystyrene surfaces. We demonstrated that straight and curved motor tracks with widths of less than 500 nm reliably guide gliding microtubules. Moreover, the performance of complex motor patterns, recently designed by evolutionary algorithms for controlling the global directionality of microtubule motion on large-area substrates, could be experimentally verified. The presented results may open up new routes toward controlling microtubule motion in self-organized hybrid-devices.
Reuther, C., Mittasch, M., Naganathan, S. R., Grill, S. W. & Diez, S. Highly-Efficient Guiding of Motile Microtubules on Non-Topographical Motor Patterns.
Nano Letters 17, 5699-5705, (2017).
doi:10.1021/acs.nanolett.7b02606