RESEARCH
RESEARCH IN THE PROJECT IS STRUCTURED INTO THE FOUR PARTS DESCRIBED BELOW.
SCIENTIFIC PUBLICATIONS FROM THE PROJECT ARE LISTED SEPARATELY
ALGORITHMS
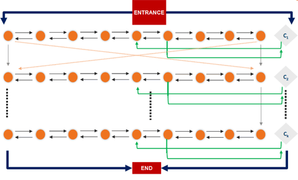
The first step in solving mathematical problems with network-based biocomputation is to encode the problem into network format so that molecular motors exploring the network can solve the problem. We have already found network encodings for several NP-complete problems, which are particularly hard to solve with electronic computers. For example, we have encoded subset sum, exact cover, boolean satisfiability and traveling salesman.
Within the Bio4Comp project, we will focus on optimizing these encodings so that they can be efficiently solved with biological agents and be more readily scaled up. Analogous to optimized computer algorithms, optimized networks can greatly reduce the computing power (and thus the number of motor proteins) required for finding the correct solution.
With larger and more complex network designs, it becomes critical, to verify that they work correctly. We will therefore develop methods and software tools to support the robust design of networks.
ARCHITECTURAL ELEMENTS
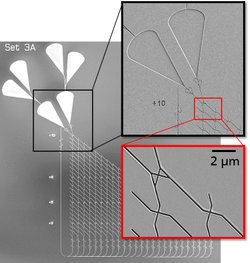
We implement network algorithms into actual physical networks by designing junctions that guide the filaments according to the rules defined by each algorithm. For example, we have designed split junctions, that equally distribute filaments to all exit channels and pass junctions that ensure that filaments always move straight across the junction. Computer simulations optimize each type of junction before fabrication and testing. We are developing new technologies such as true 3D laser writing, switchable smart polymer gates and automated detectors. These technologies will allow us to fabricate error-free 3D junctions and active junctions that can switch between different junction types. Such junctions will not only enable larger networks that can solve several different problems but also more sophisticated, dynamic programming algorithms, that can operate without human interaction.
SCALING THE BIOLOGY
The biological properties of the actin filaments and microtubules will be devised to e.g. 1. exponentially increase their number during passage of biocomputation networks and 2. store information about the paths through the network on filaments. Because actin-myosin and microtubule-kinesin motility systems have specific advantages and pose different challenges both systems are investigated. In addition to key roles for fully integrated and up-scaled biocomputation devices the work will give unique fundamental insights of value for understanding diseases where defective molecular motor and cytoskeletal systems play roles, e.g. cardiac diseases, neurodegenerative diseases and cancer.
INTEGRATION AND FABRICATION
The Bio4Comp approach needs a real scale-up from lab-scale test samples to large-scale biocomputation networks. In order to achieve both upscaling regarding network size and downscaling regarding feature dimensions, we use standard microfabrication and advanced nanopatterning techniques such as electron beam nanolithography and two-photon polymerization. Our research is devoted to process engineering and materials science to achieve the scaled-up biocomputation network structures. The work program also includes integration concepts for the various architectural components (channel junctions, agent detectors) to allow the programmability of the network structures.
"One of the most exciting aspects of network-based computing with molecular motors is that it needs hundred to thousand times less energy than electronic computers."
Heiner Linke, Project coordinator